Circular RNA
January 17, 2022 | 6 min read
January 17, 2022 | 6 min read

RNA is an exquisite biomolecule transcribed from the DNA and translated into protein a/c to the central dogma of molecular biology. It comprises the same four molecules (bases) as in DNA, except thymine. However, its ability to assume various structures deviating from Watson-Crick base pairing bestows on it hitherto unknown functions. The notion of a circular structure of RNA arose some 30 years ago from electron microscopy observations. The idea of a circular isoform of RNA produced post-transcriptionally from genes gradually developed over the next 20 years. Subsequently, circular RNAs (circRNAs) were discovered from the SRY gene, the Antisense Non-coding RNA in INK4 locus (ANRIL), Cerebellar Degeneration protein 1(CDR1). Until 2012 circRNAs were construed as anomalies or splicing noise due to their unusual structures post transcription.
The eukaryotic pre-mRNA undergoes splicing to produce mature mRNA, which is translated into proteins. The several exons in a pre-mRNA can be spliced together in different ways, called alternative splicing. In protein-coding genes, exons may be flanked by introns with reverse complementary sequence. This allows the formation of paired duplex structure where the downstream 5’ splice donor site is joined to the upstream 3’ splice acceptor site via a kind of alternative splicing called backsplicing. The resulting circRNA molecule is covalently linked by a 3’-5’ phosphodiester bond at the backspliced junction.
However, circRNA biogenesis aided by reverse complementary intronic sequences may not be sufficient but involve other processes. Exon skipping events may result in a mature, linear mRNA and an intron lariat. These may undergo resplicing to form a stable circRNA and a double lariat structure, debranched and degraded later. Recent evidence suggests that depletion or pharmacological inhibition of spliceosome proteins tend to increase circRNA levels post transcription. Spliceosome proteins U1 and U2 snRNPs assemble on different exons to form exon junction complexes. Subsequently, introns interact to form a lariat and a mature mRNA is formed. However, if spliceosome proteins are depleted or the exon is quite long, the spliceosome machinery may assemble across the same exon, thus allowing backsplicing and producing a circRNA.
The circRNAs formed are remarkably stable; their closed, circular structure prevents degradation by RNA exonucleases and consequently have half-lives > 24 hr, unlike linear mRNA. The majority of circRNAs tend to accumulate in the cytoplasm, albeit a subset is localized in the nucleus presumed to regulate transcription and splicing. Scientific evidence suggests that nuclear export of circRNA occurs in a length-dependent manner involving different set of proteins.
Although a detailed blueprint of the molecular and physiological significance of circRNA is yet to be determined, recent evidence on circRNA research exhibit functions galore. Comprehensive studies on a circRNA ciRS-7 have observed that it sequesters miR-7. Similarly, mouse testis-specific Sry circRNA has found 16 putative binding sites for miR-138. These studies propose a model wherein circRNA acts as miRNA sponge regulating their temporal and spatial localization and ensures suitable level of their delivery to miRNA target genes. However, the low level of most circRNA expression and few miRNA binding sites poses a problem and requires further investigation.
Several circRNAs have been reported to possess an internal ribosome entry site (IRES) and start codons. Further studies have suggested that circRNA may be translated into proteins. It is speculated that circRNAs may be translated in a localized manner and under specific circumstances, like stress. However, such speculation remains to be substantiated with adequate experimental proof.
Circular RNAs are abundant in eukaryotes and have been identified in hepatitis delta virus, plant viroids and other DNA viruses. This holds the problem of identification of endogenous and exogenous circRNAs i.e., self vs non-self recognition. Studies have revealed that N6-methyladenosine (m6A); one of the most abundant RNA modifications comprising 0.4-0.6% of all mRNAs, long non-coding RNAs and circRNAs, serve to distinguish between self vs non-self circRNA molecules. Recent studies have elucidated a mechanism wherein exogenous circRNAs stimulates innate immune response via RIG-1 PRRs. RIG-1 activated by unmodified exogenous circRNA induces polymerization of mitochondria anti-viral signalling protein (MAVS). MAVS filamentation leads to dimerization of transcription factor IRF-3 and allows interferon production. Circular RNA also interacts with several RNA-binding proteins; recent evidence indicates that reduced circRNA level releases ds RNA activated protein kinase (PKR), which initiates immune response on recognition of pathogenic RNAs.
It has been observed that circRNA levels are reduced during uncontrolled cell proliferation in cancerous patients. Circular RNA encoded by POKEMON gene functions as a proto-oncogene, while its linear counterpart acts as a tumour suppressor. Others like circRNA from Foxo3 gene promotes apoptosis, thus inhibits tumour growth. Elucidation of circRNA roles in tumour progression calls for further investigation.
To summarize, circRNAs are not transcription anomalies; rather they possess specific functions w.r.t transcription and splicing, gene regulation, innate immunity, cancer, to name a few. Further research is essential to completely unearth the molecular mechanisms and physiological significance of several problems discussed above.
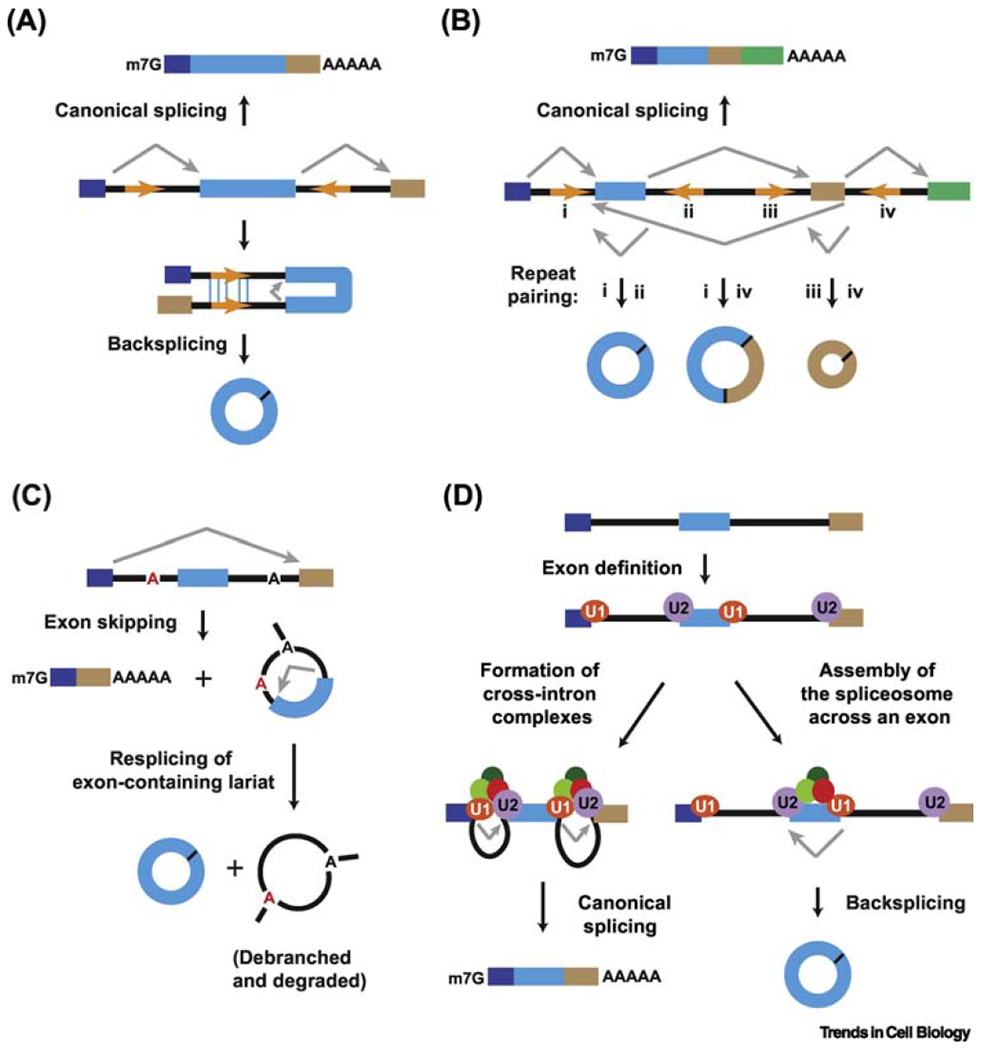
Figure 1.
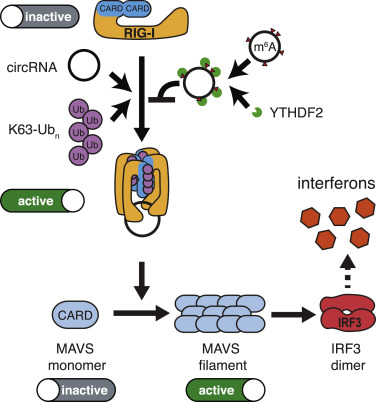
Figure 2.