RNA Nanopore
May 7, 2022 | 2 min read

May 7, 2022 | 2 min read

Viruses are one of the most rapidly mutating organisms on the planet. Their high recombination rate leads to the formation of closely related groups that are called ‘Quasispecies’, especially in RNA viruses. The variety of genomes in the total population is huge, but from individual to individual the change in the genome isn’t that large. This means that to identify different variants, sequencing parts of the genome is not enough, the complete genome of different individuals must be compared. This paper aims to categorize viral RNAs produced in cells infected with a human coronavirus by doing full-length, direct RNA sequencing (DRS) approach based on nanopores.
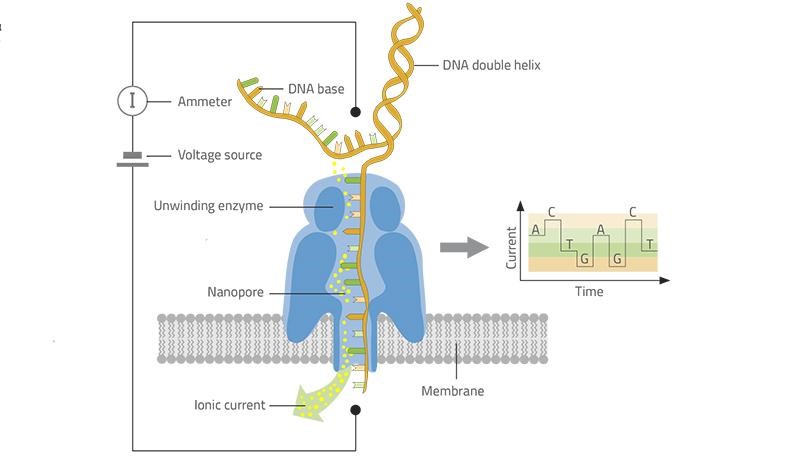
Nanopore sequencing, developed by Oxford Nanopore Technologies Ltd. is the fourth generation, scalable technology that enables direct, real-time analysis of long DNA or RNA fragments. It is a technique that does not require PCR amplification or labeling of the sample. It uses electrophoresis to transport the sample through an orifice of 10−9 m in diameter. It contains an electrolytic solution through which, when a constant electric field is applied, an electric current can be observed in the system. When close enough to nanopores, samples cause characteristic changes in electric current density across nanopore surfaces and this change is different for different nitrogen bases. The visualized raw voltage signal of a nanopore read is commonly called “squiggle”. These voltage signals are then mapped onto the bases they belong to, a process called base calling. It’s a relatively low-cost technology that has high throughput and low material requirement.
The researchers were able to directly sequence the RNA molecules of different samples of one of the largest RNA virus genomes known to date. They showed how such large RNA genomes and a diverse set of sub-genome length RNAs with complex structures can be investigated at high resolution without the need for a prior assembly step and without the bias introduced by cDNA synthesis that is typically required for transcriptome studies. Combining nanopore sequencing with illumina sequencing to cross-check the results, not only were they able to characterize the different genomes found in an infected cell, but they also identified a few novel strains of the virus. They were also able to detect methylation sites on the viral RNA as well as previously unidentified recombination sites. They also analyzed 5-methyl cytosine methylation across various RNAs and observed that there was a consistent pattern in the same genomic positions across different RNAs, suggesting that the methylation of coronavirus RNAs is sequence-specific and controlled by RNA structural elements.